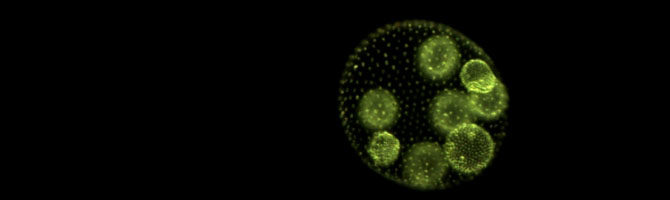
Kimberly Chen, a former postdoc in my lab, has published a new paper in Genome Biology and Evolution, “Genetic predisposition toward multicellularity in Chlamydomonas reinhardtii“:
The evolution from unicellular to multicellular organisms facilitates further phenotypic innovations, notably cellular differentiation. Multiple research groups have shown that, in the laboratory, simple, obligate multicellularity can evolve from a unicellular ancestor under appropriate selection. However, little is known about the extent to which deterministic factors such as ancestral genotype and environmental context influence the likelihood of this evolutionary transition. To test whether certain genotypes are predisposed to evolve multicellularity in different environments, we carried out a set of 24 evolution experiments, each founded by a population consisting of 10 different strains of the unicellular green alga Chlamydomonas reinhardtii, all in equal proportions. Twelve of the initially identical replicate populations were subjected to predation by the protist Paramecium tetraurelia, while the other 12 were subjected to settling selection by slow centrifugation. Population subsamples were transferred to fresh media on a weekly basis for a total of 40 transfers (∼600 generations). Heritable multicellular structures arose in 4 of 12 predation-selected populations (6 multicellular isolates in total), but never in the settling selection populations. By comparing whole genome sequences of the founder and evolved strains, we discovered that every multicellular isolate arose from one of two founders. Cell cluster size varied not only among evolved strains derived from different ancestors but also among strains derived from the same ancestor. These findings show that both deterministic and stochastic factors influence whether initially unicellular populations can evolve simple multicellular structures.
I Chen Kimberly Chen, Shania Khatri, Matthew D Herron, Frank Rosenzweig, Genetic Predisposition Toward Multicellularity in Chlamydomonas reinhardtii, Genome Biology and Evolution, 17:evaf090, https://doi.org/10.1093/gbe/evaf090